What Was The Size Of Your Amplified Dna Sample?
Agarose gel electrophoresis separates biomolecules, such equally DNA and proteins, into discrete bands each comprised of the same sized molecules. By separating these important molecules past size scientists can isolate, identify and/or clarify them. [For a fun and fast 101 on electrophoresis cheque out the Amoeba sister's video and for a deeper dive – and to see a existent electrophoresis experiment in action – view our recent live stream episode.]
Often the results from an electrophoresis experiment can exist analyzed by straight comparing the bands produced by an experiment'south samples to those produced by several controls. For instance, in the gel beneath the start three lanes are controls that prove the Dna banding patterns produced when a person has sickle cell affliction (lane 1), is a carrier with a single sickle-cell gene (lane 2), and is healthy and has 2 normal genes (lane 3). In this instance, determining the genotype of the 3 patients on the left (lanes four, v, and half dozen) at the sickle cell gene can be achieved past matching the banding blueprint of their DNA to one of the controls.

Nevertheless, at other times the actual length of a fragment is needed. Because the building blocks of Dna are nucleotides, Deoxyribonucleic acid is measured in nucleotide base of operations pairs or bp for short. Knowing the bp length of a Deoxyribonucleic acid fragment tin be essential when working with repetitive Deoxyribonucleic acid regions like microsatellites, when constructing recombinant DNA plasmids, or when collecting information for big databases. In many cases, length tin also provide added evolutionary information.
To determine a DNA molecule's base pair length a particular type of control is needed called a Dna Standard Mark or sometimes a Deoxyribonucleic acid Ladder. This is a sample that contains a mixture of Dna fragments whose lengths are already known. These fragments are used to create a standard curve that describes the human relationship between Dna size and altitude traveled during an experiment. The equation derived from this bend tin can and then be used to calculate the length of whatever unknown fragments based on how far these fragments traveled during the same experiment.
What's log got to do with it?
Larger molecules travel slowly through an agarose gel while smaller molecules travel apace. This ways that the migrate rate of a fragment is inversely proportional to the length of the molecule.
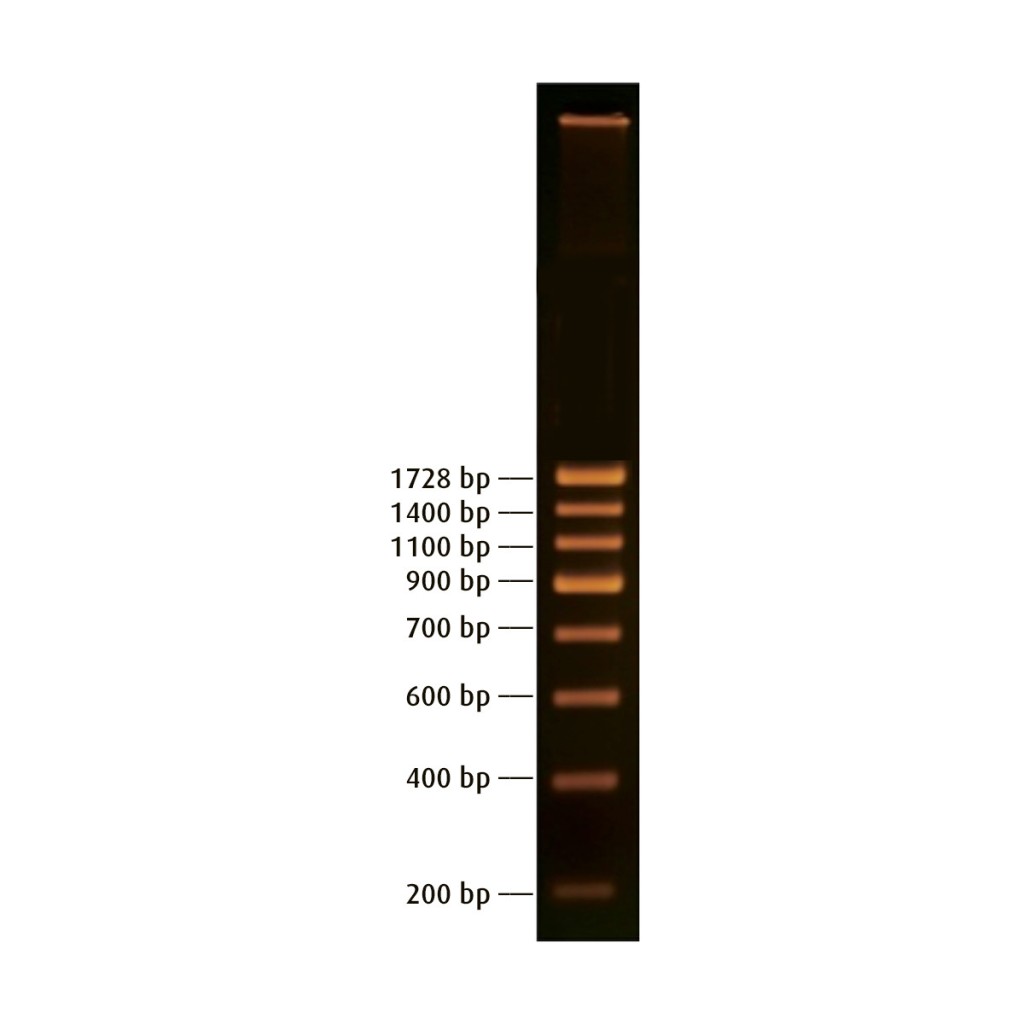
Dna Ladder with larger / slower moving fragments well-nigh the top and smaller / faster moving fragments near the bottom.
However, this relationship is not linear. Rather as fragments get smaller they move faster and faster and as they become larger they move slower and slower. For instance, in the DNA ladder in a higher place the base pair deviation between the top two fragments is 328 bp while the difference between the bottom two fragments is 200 bp. Even so there is more distance betwixt the bottom two fragments because these smaller fragments traveled much faster through the gel.
You can observe this inverse only non-linear when y'all create a graph of known fragment lengths versus distance traveled as I've done beneath for Edvotek'south Standard Marker (a picture of the this ladder is at the end of the post). In this graph the distances each fragment traveled are on the 10-axis and the bp lengths of the fragments are on the y-axis. This graph was created in a spreadsheet as was the equation that'due south displayed in the right-manus corner.

Another way to plot this information is in a log graph which is shown below. Like the first graph the distances each fragment traveled are on the x-centrality and the bp length of the fragments are on the y-axis. However, unlike the previous graph, the y-axis is on a log calibration. Fifty-fifty though these two graphs look different they use the aforementioned information and describe the same relationship. A log scale graph is merely a tool we can use to run across the information differently. Importantly, a log scale graph is also much easier to create and use if you need to calculate bp lengths by hand because its easier to draw and calculate the gradient when the graph'southward trendline is a straight line.

Sometimes logs can trip us up. Only they are really just a mode to write exponential equations so that the exponent is isolated on one side of the equation – e.g. 23 = 8 can exist written as logii 8 = 3. In fact, logs were invented to make life easier for scientists working with very big numbers. The method was originally adult in the early 1600s by John Napier and gained popularity amongst scientists of the day because it helped them to more accurately and hands calculate planetary orbits. Today, logs are still used every bit a manner to effectively draw datasets where there is a large range of number values. Google uses logs to rank pages in its search engine and many economists use logs to describe countries economic growth rates. In electrophoresis, logs make nucleotide base pair measurements that can vary from 100 to 12,000 in the space of a few centimeters easier to graph and analyze.
Ready, fix, run! (Then measure, graph, and calculate!)
Follow the steps beneath to discover the lengths of different DNA molecules. If you don't have a gel on hand you lot can use the prototype at the bottom of this post to practice.
Stride 0: Run a ladder in your gel. This sample will contain bands of known length which tin be used to create a standard curve. Every experimental run of electrophoresis has its own unique gear up of weather so every gel should take a ladder.
Stride ane: Measure and record the distance traveled by each band in the ladder and record this distance in your notebook. Traditionally, this measurement is taken from the lower edge of the sample well to the lower edge of each band.
Step 2: Repeat stride i for the fragments of the unknown samples.
Stride 3: Generate a standard curve past plotting the data on a semi-log plot. (Yous can make semilog graph paper past going here and so printing page xvi.) To create a standard bend on the semi-log paper, plot the distance each Standard Marker DNA fragment migrated on the x-axis (in mm) versus its size on the y-centrality (in base pairs). Be sure to label the axes! After all the points have been plotted, use a ruler or a directly edge to draw the best directly line possible through the points. The line should have a similar number of points on each side of the line. It's okay if the line runs through some points.

Step iv: Determine the length of each unknown fragment. Commencement, locate the migration distance of the unknown fragment on the x-axis of the graph paper. Next, draw a vertical line from this point until it intersects the line of your standard curve. Finally, from this bespeak of intersection draw a 2d line, this time horizontally, towards the y-axis. The value at which this line intersects the y-axis represents the approximate size of the fragment in base of operations pairs.
This is simply one mode of doing this. There are many dissimilar approaches yous tin can utilize. For instance, steps three and 4 tin can be done in excel or a similar spreadsheet software by using the plan's graphing and trendline functions and and so plugging the stride ii measurements into the resulting equation. Another opinion (if you have a ladder with <6 bands) is to become here and simply enter in the altitude measurements (the same calculations are being done but backside the scenes). Finally, the costless software programme Factor Clarify or the popular schoolhouse data program Logger Pro tin can be used to complete all 5 steps.

Happy molecule measuring!
What Was The Size Of Your Amplified Dna Sample?,
Source: https://blog.edvotek.com/2020/05/26/biotech-basics-sizing-dna-fragments/
Posted by: doylecriall97.blogspot.com


0 Response to "What Was The Size Of Your Amplified Dna Sample?"
Post a Comment